Chapter 25 Managing Sequence Metadata
25.1 What is Sequence Metadata?
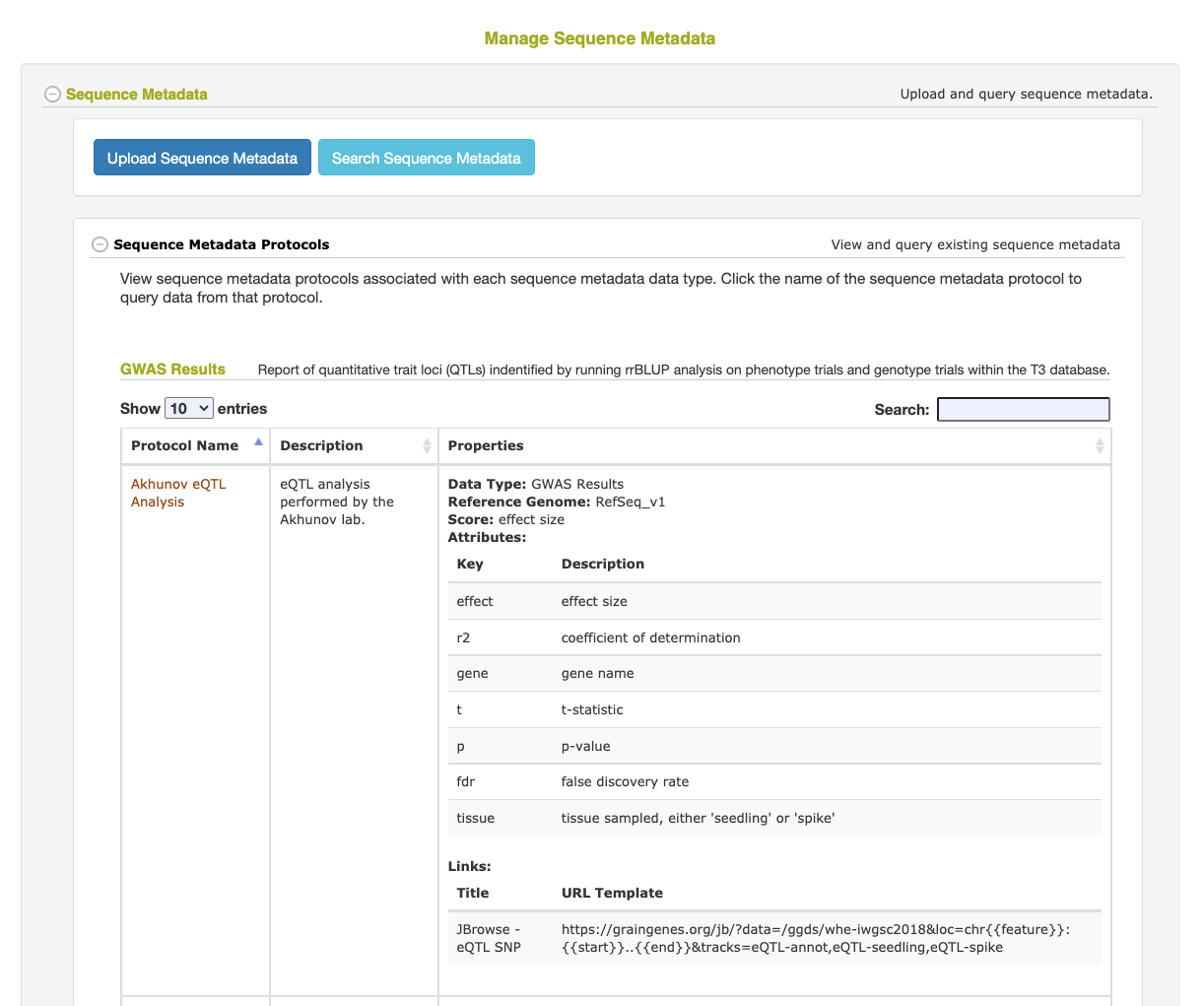
Sequence Metadata is a feature that allows for the efficient storage and retrieval of sequence annotations for a specific region along a reference genome. The annotation data can contain a primary “score” value and any number of secondary key/value attribute data. For example, Sequence Metatadata can store MNase open chromatin scores for every 10 basepairs along the reference genome as well as genome-wide association study (GWAS) statistics, including the trait information associated with the result. This data can then be filtered by position and/or scores/attribute values and even cross-referenced with markers stored in the database.
25.2 Loading Sequence Metadata
Sequence Metadata can be loaded into the database using a gff3-formatted file. The following columns are used to load the data:
- #1 / seqid: The name of the database feature (ie chromosome) the metadata is associated with (The feature name must already exist as a feature in the database)
- #4 / start: The metadata’s start position
- #5 / end: The metadata’s end position
- #6 / score: (optional) The primary score attribute of the metadata
- #9 / attributes: (optional) Secondary key//value attributes to be saved with the score. These should be formatted using the gff3 standard (key1=value1;key2=value2). The attribute key cannot be either score, start, or end.
To upload the gff3 file:
- Go to the Manage > Sequence Metadata page
- Click the Upload Sequence Metadata button
- On Step 2 of the Wizard, select the Type of data to be uploaded
- This groups similar datasets together in the same Data Type category
- On Step 3 of the Wizard, select an existing Protocol or create a new one
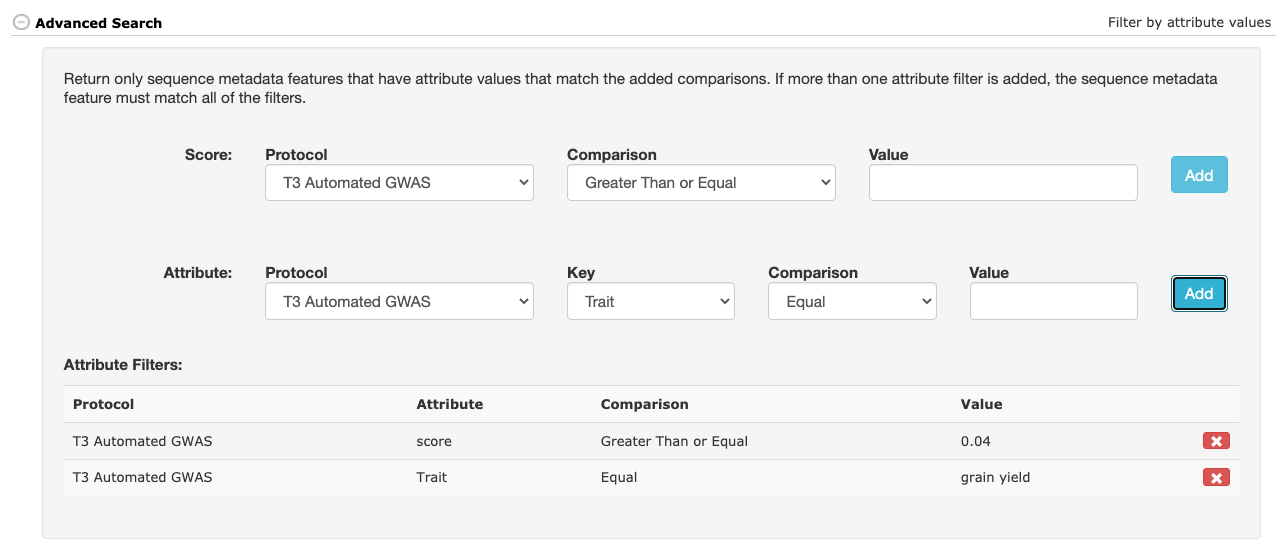
- The Protocol is used to describe how the data was generated and define the score value and any secondary attributes. Adding the attributes (and their descriptions) to the Protocol will allow the Sequence Metadata queries to filter the data based on the value of one or more of these attributes. Attributes not defined in the Protocol will still be stored and displayed on retrieval, but will not be able to be used in a search filter.
- Finally, select and upload your gff3 file to the database. The database will verify the format of the file before its contents are stored.
25.3 Searching Sequence Metadata
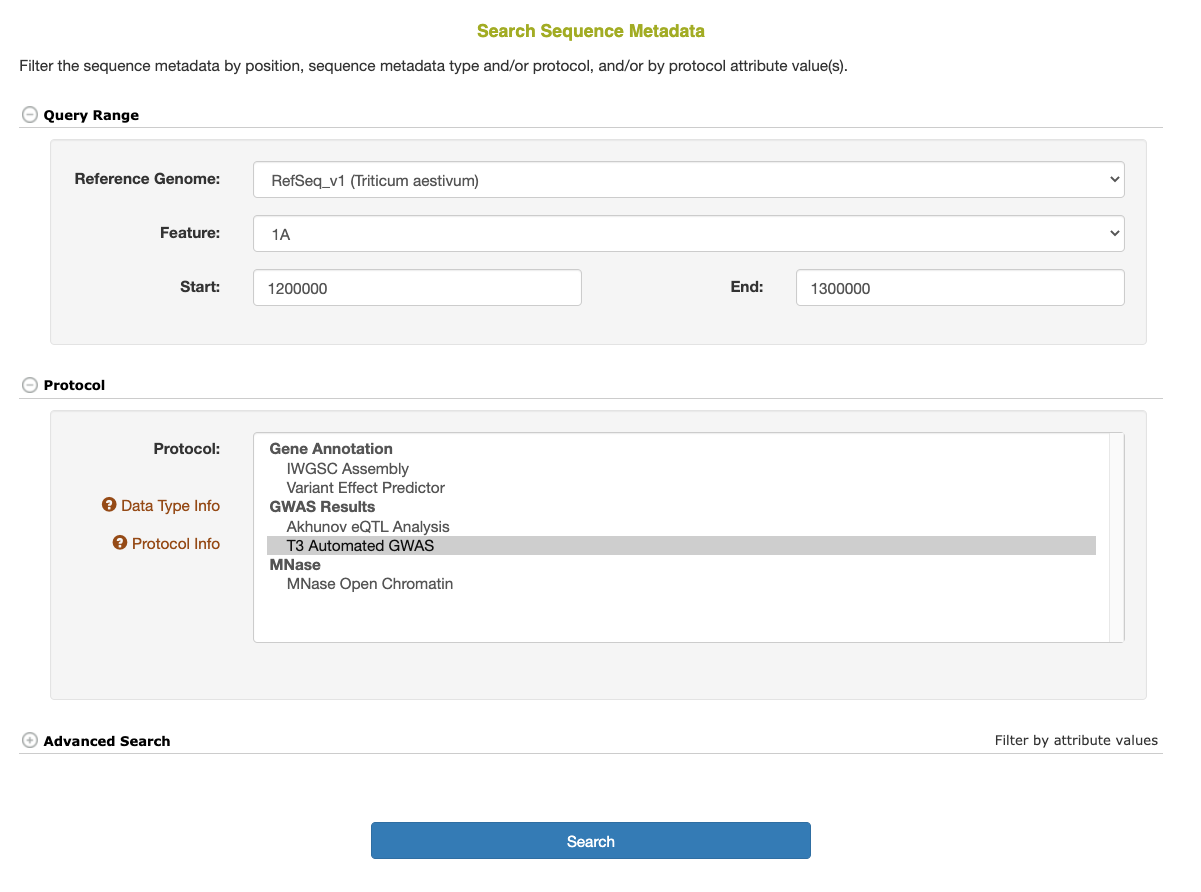
To retrieve stored Sequence Metadata, go to the Search > Sequence Metadata page.
25.3.1 Basic Search
The basic Sequence Metadata search options include selecting the reference genome and species, the chromosome, and (optionally) the start and/or end position(s) along the reference genome. In addition, one or more specific protocols can be selected to limit the results.
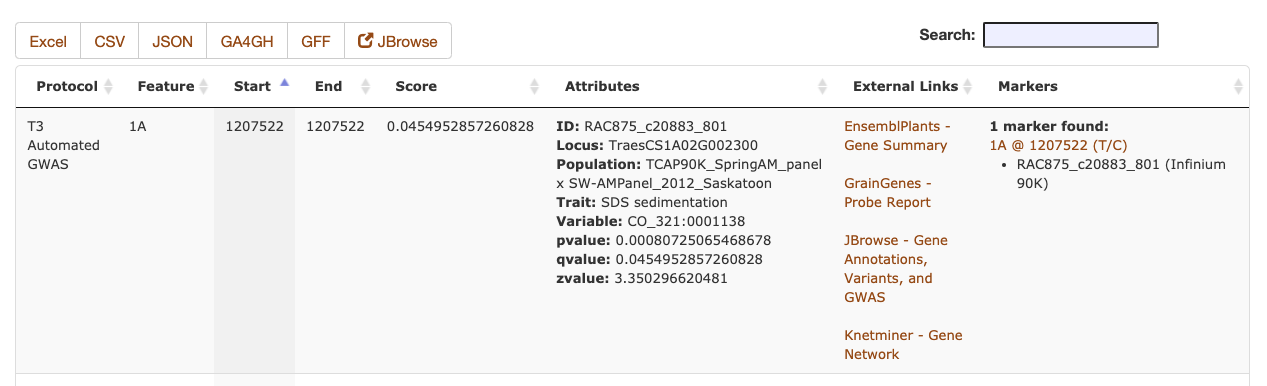
The Sequence Metadata search results are returned as a table, including the chromosome and start/stop positions of the annotation, along with the primary score value and any additional key/value attributes. The markers column will include a list of marker names of any stored markers that are found within the start/stop positions of the Sequence Metadata. The data can be downloaded as a table in an Excel or CSV file or a machine-readable (code-friendly) JSON file. If the Sequence Metadata JBrowse configuration is set, the filtered results can be displayed as a dynamic JBrowse track.
25.4 Marker Integration
A table of Sequence Metadata annotations are embedded on the Marker/Variant detail page. The table will include any annotations that span the poisiton of the marker (for data of the same reference genome and species).
25.5 Sequence Metadata API
A publicly accessible RESTful API (Application Programming Interface) is available to query the database for Sequence Metadata directly from your programming environment (R, python, etc) to be used in analysis. The data is returned in a JSON format. Documentation for the API can be found on the Manage > Sequence Metadata page